



Statistica Sinica 12(2002), 241-262
EVALUATION AND COMPARISON OF
CLUSTERING ALGORITHMS IN ANGLYZING ES
CELL GENE EXPRESSION DATA
Gengxin Chen
,
Saied A. Jaradat
,
Nila Banerjee
,
Tetsuya S. Tanaka
,
Minoru S. H. Ko
and Michael Q.
Zhang
 Cold Spring Harbor Laboratory and
Cold Spring Harbor Laboratory and
 National Institutes of Health, U.S.A.
National Institutes of Health, U.S.A.
Abstract:
Many clustering algorithms have been used to analyze microarray gene
expression data.
Given embryonic stem cell gene expression data,
we applied several indices to evaluate
the performance of clustering algorithms,
including hierarchical clustering, 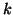 -means,
PAM and SOM. The indices were homogeneity and separation scores,
silhouette width,
redundant score (based on redundant genes), and WADP
(testing the robustness of
clustering results after small perturbation).
The results showed that the ES cell dataset
posed a challenge for cluster analysis in that the clusters generated
by different methods were only partially consistent.
Using this data set, we were able to evaluate the advantages and
weaknesses of algorithms with respect to both internal and external
quality measures.
This study may provide a guideline on how to select suitable clustering
algorithms and it may help raise issues in the extraction of
meaningful biological information from microarray expression data.
-means,
PAM and SOM. The indices were homogeneity and separation scores,
silhouette width,
redundant score (based on redundant genes), and WADP
(testing the robustness of
clustering results after small perturbation).
The results showed that the ES cell dataset
posed a challenge for cluster analysis in that the clusters generated
by different methods were only partially consistent.
Using this data set, we were able to evaluate the advantages and
weaknesses of algorithms with respect to both internal and external
quality measures.
This study may provide a guideline on how to select suitable clustering
algorithms and it may help raise issues in the extraction of
meaningful biological information from microarray expression data.
Key words and phrases:
Cluster analysis, gene expression, microarray, mouse embryonic stem cell.








-means, PAM and SOM. The indices were homogeneity and separation scores, silhouette width, redundant score (based on redundant genes), and WADP (testing the robustness of clustering results after small perturbation). The results showed that the ES cell dataset posed a challenge for cluster analysis in that the clusters generated by different methods were only partially consistent. Using this data set, we were able to evaluate the advantages and weaknesses of algorithms with respect to both internal and external quality measures. This study may provide a guideline on how to select suitable clustering algorithms and it may help raise issues in the extraction of meaningful biological information from microarray expression data.



